数学建模学习笔记(二十)TSP问题遗传算法求解
【摘要】
什么是TSP问题? TSP问题(Travelling Salesman Problem)又译为旅行推销员问题、货郎担问题,即假设有一个旅行商人要拜访n个城市,从某个城市出发,每个城市只能访问一次且最后回到...
什么是TSP问题?
TSP问题(Travelling Salesman Problem)又译为旅行推销员问题、货郎担问题,即假设有一个旅行商人要拜访n个城市,从某个城市出发,每个城市只能访问一次且最后回到出发城市,问该推销员应如何选择路线,才能使总的行程最短?
使用遗传算法求解:
代码转于知乎博主https://zhuanlan.zhihu.com/p/153098599
from math import floor
import numpy as np
import matplotlib.pyplot as plt # 导入所需要的库
class Gena_TSP(object):
def __init__(self, data, maxgen=200, size_pop=200, cross_prob=0.9, pmuta_prob=0.01, select_prob=0.8):
self.maxgen = maxgen # 最大迭代次数
self.size_pop = size_pop # 群体个数
self.cross_prob = cross_prob # 交叉概率
self.pmuta_prob = pmuta_prob # 变异概率
self.select_prob = select_prob # 选择概率
self.data = data # 城市的左边数据
self.num = len(data) # 城市个数 对应染色体长度
self.matrix_distance = self.matrix_dis()
# 距离矩阵n*n, 第[i,j]个元素表示城市i到j距离matrix_dis函数见下文
self.select_num = max(floor(self.size_pop * self.select_prob + 0.5), 2)
# 通过选择概率确定子代的选择个数
self.chrom = np.array([0] * self.size_pop * self.num).reshape(self.size_pop, self.num)
self.sub_sel = np.array([0] * self.select_num * self.num).reshape(self.select_num, self.num)
# 父代和子代群体的初始化(不直接用np.zeros是为了保证单个染色体的编码为整数,np.zeros对应的数据类型为浮点型)
self.fitness = np.zeros(self.size_pop)
# 存储群体中每个染色体的路径总长度,对应单个染色体的适应度就是其倒数
self.best_fit = []
self.best_path = []
# 保存每一步的群体的最优路径和距离
def matrix_dis(self):
res = np.zeros((self.num, self.num))
for i in range(self.num):
for j in range(i + 1, self.num):
res[i, j] = np.linalg.norm(self.data[i, :] - self.data[j, :])
res[j, i] = res[i, j]
return res
def rand_chrom(self):
rand_ch = np.array(range(self.num))
for i in range(self.size_pop):
np.random.shuffle(rand_ch)
self.chrom[i,:]= rand_ch
self.fitness[i] = self.comp_fit(rand_ch)
def comp_fit(self, one_path):
res = 0
for i in range(self.num - 1):
res += self.matrix_distance[one_path[i], one_path[i + 1]]
res += self.matrix_distance[one_path[-1], one_path[0]]
return res
def out_path(self, one_path):
res = str(one_path[0] + 1) + '-->'
for i in range(1, self.num):
res += str(one_path[i] + 1) + '-->'
res += str(one_path[0] + 1) + '\n'
print(res)
def select_sub(self):
fit = 1. / (self.fitness) # 适应度函数
cumsum_fit = np.cumsum(fit)
pick = cumsum_fit[-1] / self.select_num * (np.random.rand() + np.array(range(self.select_num)))
i, j = 0, 0
index = []
while i < self.size_pop and j < self.select_num:
if cumsum_fit[i] >= pick[j]:
index.append(i)
j += 1
else:
i += 1
self.sub_sel = self.chrom[index, :]
def cross_sub(self):
if self.select_num % 2 == 0:
num = range(0, self.select_num, 2)
else:
num = range(0, self.select_num - 1, 2)
for i in num:
if self.cross_prob >= np.random.rand():
self.sub_sel[i, :], self.sub_sel[i + 1, :] = self.intercross(self.sub_sel[i, :], self.sub_sel[i + 1, :])
def intercross(self, ind_a, ind_b):
r1 = np.random.randint(self.num)
r2 = np.random.randint(self.num)
while r2 == r1:
r2 = np.random.randint(self.num)
left, right = min(r1, r2), max(r1, r2)
ind_a1 = ind_a.copy()
ind_b1 = ind_b.copy()
for i in range(left, right + 1):
ind_a2 = ind_a.copy()
ind_b2 = ind_b.copy()
ind_a[i] = ind_b1[i]
ind_b[i] = ind_a1[i]
x = np.argwhere(ind_a == ind_a[i])
y = np.argwhere(ind_b == ind_b[i])
if len(x) == 2:
ind_a[x[x != i]] = ind_a2[i]
if len(y) == 2:
ind_b[y[y != i]] = ind_b2[i]
return ind_a, ind_b
def mutation_sub(self):
for i in range(self.select_num):
if np.random.rand() <= self.cross_prob:
r1 = np.random.randint(self.num)
r2 = np.random.randint(self.num)
while r2 == r1:
r2 = np.random.randint(self.num)
self.sub_sel[i, [r1, r2]] = self.sub_sel[i, [r2, r1]]
def reverse_sub(self):
for i in range(self.select_num):
r1 = np.random.randint(self.num)
r2 = np.random.randint(self.num)
while r2 == r1:
r2 = np.random.randint(self.num)
left, right = min(r1, r2), max(r1, r2)
sel = self.sub_sel[i, :].copy()
sel[left:right + 1] = self.sub_sel[i, left:right + 1][::-1]
if self.comp_fit(sel) < self.comp_fit(self.sub_sel[i, :]):
self.sub_sel[i, :] = sel
def reins(self):
index = np.argsort(self.fitness)[::-1]
self.chrom[index[:self.select_num], :] = self.sub_sel
def main(data):
Path_short = Gena_TSP(data) # 根据位置坐标,生成一个遗传算法类
Path_short.rand_chrom() # 初始化父类
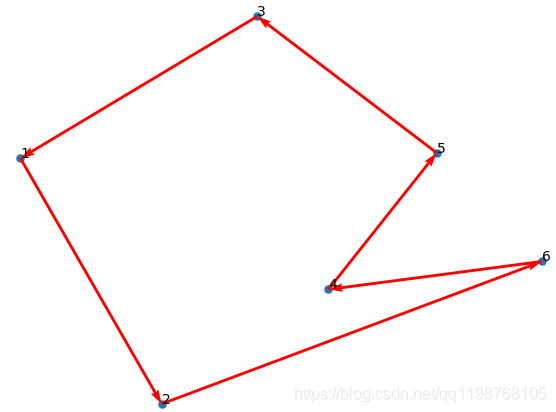
## 绘制初始化的路径图
fig, ax = plt.subplots()
x = data[:, 0]
y = data[:, 1]
ax.scatter(x, y, linewidths=0.1)
for i, txt in enumerate(range(1, len(data) + 1)):
ax.annotate(txt, (x[i], y[i]))
res0 = Path_short.chrom[0]
x0 = x[res0]
y0 = y[res0]
for i in range(len(data) - 1):
plt.quiver(x0[i], y0[i], x0[i + 1] - x0[i], y0[i + 1] - y0[i], color='r', width=0.005, angles='xy', scale=1,
scale_units='xy')
plt.quiver(x0[-1], y0[-1], x0[0] - x0[-1], y0[0] - y0[-1], color='r', width=0.005, angles='xy', scale=1,
scale_units='xy')
plt.show()
print('初始染色体的路程: ' + str(Path_short.fitness[0]))
# 循环迭代遗传过程
for i in range(Path_short.maxgen):
Path_short.select_sub() # 选择子代
Path_short.cross_sub() # 交叉
Path_short.mutation_sub() # 变异
Path_short.reverse_sub() # 进化逆转
Path_short.reins() # 子代插入
# 重新计算新群体的距离值
for j in range(Path_short.size_pop):
Path_short.fitness[j] = Path_short.comp_fit(Path_short.chrom[j, :])
# 每隔三十步显示当前群体的最优路径
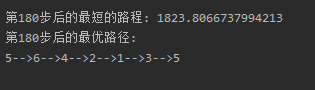
index = Path_short.fitness.argmin()
if (i + 1) % 30 == 0:
print('第' + str(i + 1) + '步后的最短的路程: ' + str(Path_short.fitness[index]))
print('第' + str(i + 1) + '步后的最优路径:')
Path_short.out_path(Path_short.chrom[index, :]) # 显示每一步的最优路径
# 存储每一步的最优路径及距离
Path_short.best_fit.append(Path_short.fitness[index])
Path_short.best_path.append(Path_short.chrom[index, :])
return Path_short # 返回遗传算法结果类
data = np.array([149,663,170,511,303,287,404,707,520,490]).reshape(5,2)
main(data)
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
- 53
- 54
- 55
- 56
- 57
- 58
- 59
- 60
- 61
- 62
- 63
- 64
- 65
- 66
- 67
- 68
- 69
- 70
- 71
- 72
- 73
- 74
- 75
- 76
- 77
- 78
- 79
- 80
- 81
- 82
- 83
- 84
- 85
- 86
- 87
- 88
- 89
- 90
- 91
- 92
- 93
- 94
- 95
- 96
- 97
- 98
- 99
- 100
- 101
- 102
- 103
- 104
- 105
- 106
- 107
- 108
- 109
- 110
- 111
- 112
- 113
- 114
- 115
- 116
- 117
- 118
- 119
- 120
- 121
- 122
- 123
- 124
- 125
- 126
- 127
- 128
- 129
- 130
- 131
- 132
- 133
- 134
- 135
- 136
- 137
- 138
- 139
- 140
- 141
- 142
- 143
- 144
- 145
- 146
- 147
- 148
- 149
- 150
- 151
- 152
- 153
- 154
- 155
- 156
- 157
- 158
- 159
- 160
- 161
- 162
- 163
- 164
- 165
- 166
- 167
- 168
- 169
- 170
- 171
- 172
- 173
- 174
- 175
- 176
- 177
- 178
- 179
遗传算法尚未看懂,先用再说,运行结果如下:
文章来源: zstar.blog.csdn.net,作者:zstar-_,版权归原作者所有,如需转载,请联系作者。
原文链接:zstar.blog.csdn.net/article/details/113481538
【版权声明】本文为华为云社区用户转载文章,如果您发现本社区中有涉嫌抄袭的内容,欢迎发送邮件进行举报,并提供相关证据,一经查实,本社区将立刻删除涉嫌侵权内容,举报邮箱:
cloudbbs@huaweicloud.com
- 点赞
- 收藏
- 关注作者

评论(0)