【图像融合】基于matlab对比度和结构提取多模态解剖图像融合【含Matlab源码 1539期】
【摘要】
一、图像融合简介
应用多模态图像的配准与融合技术,可以把不同状态的医学图像有机地结合起来,为临床诊断和治疗提供更丰富的信息。介绍了多模态医学图像配准与融合的概念、方法及意义。最后简单介绍了小波变换分析方...
一、图像融合简介
应用多模态图像的配准与融合技术,可以把不同状态的医学图像有机地结合起来,为临床诊断和治疗提供更丰富的信息。介绍了多模态医学图像配准与融合的概念、方法及意义。最后简单介绍了小波变换分析方法。
二、部分源代码
clear; close all; clc; warning off
%% A Novel Multi-Modality Anatomical Image FusionMethod Based on Contrast and Structure Extraction
% F = fuseImage(I,scale)
%Inputs:
%I - a mulyi-modal anatomical image sequence
%scale - scale factor of dense SIFT, the default value is 16
%% load images from the folder that contain multi-modal image to be fused
%I=load_images('./Dataset\CT-MRI\Pair 1');
I=load_images('./Dataset\MR-T1-MR-T2\Pair 1');
%I=load_images('./Dataset\MR-Gad-MR-T1\Pair 1');
% Show source input images
figure;
no_of_images = size(I,4);
for i = 1:no_of_images
subplot(2,1,i); imshow(I(:,:,:,i));
end
suptitle('Source Images');
%%
F=fuseImage(I,16);
%% Output: F - the fused image
F=rgb2gray(F);
figure;
imshow(F);
function [ F ] = fuseImage(I,scale)
addpath('Pyramid_Decomposition');
addpath('Guided_Filter');
addpath('Dense_SIFT');
tic
%%
[H, W, C, N]=size(I);
imgs=im2double(I);
IA=zeros(H,W,C,N);
for i=1:N
IA(:,:,:,i)=enhnc(imgs(:,:,:,i));
end
%%
imgs_gray=zeros(H,W,N);
for i=1:N
imgs_gray(:,:,i)=rgb2gray(IA(:,:,:,i));
end
%
% %dense sift calculation
dsifts=zeros(H,W,32,N, 'single');
for i=1:N
img=imgs_gray(:,:,i);
ext_img=img_extend(img,scale/2-1);
[dsifts(:,:,:,i)] = DenseSIFT(ext_img, scale, 1);
end
%%
%local contrast
contrast_map=zeros(H,W,N);
for i=1:N
contrast_map(:,:,i)=sum(dsifts(:,:,:,i),3);
end
%winner-take-all weighted average strategy for local contrast
[x, labels]=max(contrast_map,[],3);
clear x;
for i=1:N
mono=zeros(H,W);
mono(labels==i)=1;
contrast_map(:,:,i)=mono;
end
%% Structure
h = [1 -1];
structure_map=zeros(H,W,N);
for i=1:N
structure_map(:,:,i) = abs(conv2(imgs_gray(:,:,i),h,'same')) + abs(conv2(imgs_gray(:,:,i),h','same')); %EQ 13
end
%winner-take-all weighted average strategy for structure
[a, label]=max(structure_map,[],3);
clear x;
for i=1:N
monoo=zeros(H,W);
monoo(label==i)=1;
structure_map(:,:,i)=monoo;
end
%%
weight_map=structure_map.*contrast_map;
%weight map refinement using Guided Filter
for i=1:N
weight_map(:,:,i) = fastGF(weight_map(:,:,i),12,0.25,2.5);
end
% normalizing weight maps
%
weight_map = weight_map + 10^-25; %avoids division by zero
weight_map = weight_map./repmat(sum(weight_map,3),[1 1 N]);
%% Pyramid Decomposition
% create empty pyramid
pyr = gaussian_pyramid(zeros(H,W,3));
nlev = length(pyr);
% multiresolution blending
for i = 1:N
% construct pyramid from each input image
% blend
for b = 1:nlev
w = repmat(pyrW{b},[1 1 3]);
pyr{b} = pyr{b} + w .*pyrI{b};
end
end
% reconstruct
F = reconstruct_laplacian_pyramid(pyr);
toc
end
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
- 53
- 54
- 55
- 56
- 57
- 58
- 59
- 60
- 61
- 62
- 63
- 64
- 65
- 66
- 67
- 68
- 69
- 70
- 71
- 72
- 73
- 74
- 75
- 76
- 77
- 78
- 79
- 80
- 81
- 82
- 83
- 84
- 85
- 86
- 87
- 88
- 89
- 90
- 91
- 92
- 93
- 94
- 95
- 96
- 97
- 98
- 99
- 100
- 101
- 102
- 103
- 104
- 105
- 106
- 107
- 108
- 109
- 110
- 111
- 112
- 113
- 114
- 115
- 116
- 117
- 118
- 119
- 120
- 121
- 122
- 123
- 124
- 125
- 126
- 127
- 128
- 129
- 130
- 131
- 132
- 133
- 134
- 135
- 136
- 137
- 138
- 139
- 140
- 141
- 142
- 143
- 144
- 145
- 146
- 147
- 148
- 149
- 150
- 151
三、运行结果

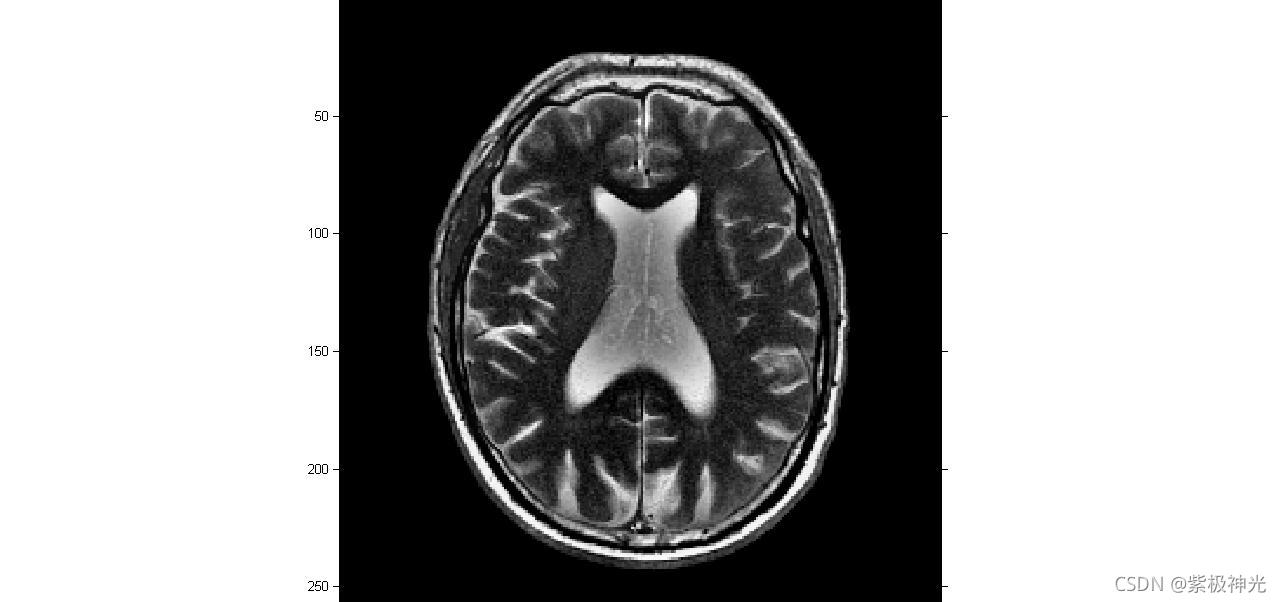
四、matlab版本及参考文献
1 matlab版本
2014a
2 参考文献
[1] 蔡利梅.MATLAB图像处理——理论、算法与实例分析[M].清华大学出版社,2020.
[2]杨丹,赵海滨,龙哲.MATLAB图像处理实例详解[M].清华大学出版社,2013.
[3]周品.MATLAB图像处理与图形用户界面设计[M].清华大学出版社,2013.
[4]刘成龙.精通MATLAB图像处理[M].清华大学出版社,2015.
[5]朱俊林.浅析多模态医学图像的配准与融合技术[J].医疗卫生装备. 2005,(12)
文章来源: qq912100926.blog.csdn.net,作者:海神之光,版权归原作者所有,如需转载,请联系作者。
原文链接:qq912100926.blog.csdn.net/article/details/121590311
【版权声明】本文为华为云社区用户转载文章,如果您发现本社区中有涉嫌抄袭的内容,欢迎发送邮件进行举报,并提供相关证据,一经查实,本社区将立刻删除涉嫌侵权内容,举报邮箱:
cloudbbs@huaweicloud.com
- 点赞
- 收藏
- 关注作者

评论(0)