基于RDKit的溶解度预测的机器学习模型
【摘要】
基于RDKit和Python3的化合物溶解度的机器学习模型小案例。
代码示例(仅供参考):
# In[1]:导入依赖包from rdkit import Chem, DataStructsfrom rdkit.Chem import AllChemfrom rdkit.ML.Descriptors import MoleculeDescriptorsfrom rdk...
基于RDKit和Python3的化合物溶解度的机器学习模型小案例。
代码示例(仅供参考):
-
# In[1]:导入依赖包
-
from rdkit import Chem, DataStructs
-
from rdkit.Chem import AllChem
-
from rdkit.ML.Descriptors import MoleculeDescriptors
-
from rdkit.Chem import Descriptors
-
from rdkit.Chem.EState import Fingerprinter
-
import pandas as pd
-
import numpy as np
-
import matplotlib.pyplot as plt
-
from sklearn.preprocessing import StandardScaler
-
from sklearn import cross_validation
-
from sklearn.metrics import r2_score
-
from sklearn.ensemble import RandomForestRegressor
-
from sklearn import gaussian_process
-
from sklearn.gaussian_process.kernels import Matern, WhiteKernel, ConstantKernel, RBF
-
# In[2]:定义描述符计算函数
-
def get_fps(mol):
-
calc=MoleculeDescriptors.MolecularDescriptorCalculator([x[0] for x in Descriptors._descList])
-
ds = np.asarray(calc.CalcDescriptors(mol))
-
arr=Fingerprinter.FingerprintMol(mol)[0]
-
return np.append(arr,ds)
-
# In[3]:
-
#读入数据
-
data = pd.read_table('smi_sol.dat', sep=' ')
-
#增加结构和描述符属性
-
data['Mol'] = data['smiles'].apply(Chem.MolFromSmiles)
-
data['Descriptors'] = data['Mol'].apply(get_fps)
-
# In[4]:查看前5行数据
-
data.head(5)
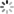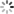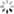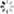
-
# In[5]:
-
#转换为numpy数组
-
X = np.array(list(data['Descriptors']))
-
y = data['solubility'].values
-
st = StandardScaler()
-
X = st.fit_transform(X)
-
#划分训练集和测试集
-
X_train, X_test, y_train, y_test = cross_validation.train_test_split(X, y, test_size=0.25, random_state=42)
-
# In[7]:高斯过程回归
-
kernel=1.0 * RBF(length_scale=1) + WhiteKernel(noise_level=1)
-
gp = gaussian_process.GaussianProcessRegressor(kernel=kernel,n_restarts_optimizer=0,normalize_y=True)
-
gp.fit(X_train, y_train)
-
# In[8]:
-
y_pred, sigma = gp.predict(X_test, return_std=True)
-
rms = (np.mean((y_test - y_pred)**2))**0.5
-
print ("GP RMS", rms)
-
# out[8]:
-
GP RMS 0.5984083408596741
-
# In[9]:
-
print ("GP r^2 score",r2_score(y_test,y_pred))
-
# out[8]:
-
GP r^2 score 0.9141780584554846
-
# In[10]:结果绘图
-
plt.scatter(y_train,gp.predict(X_train), label = 'Train', c='blue')
-
plt.title('GP Predictor')
-
plt.xlabel('Measured Solubility')
-
plt.ylabel('Predicted Solubility')
-
plt.scatter(y_test,gp.predict(X_test),c='lightgreen', label='Test', alpha = 0.8)
-
plt.legend(loc=4)
-
plt.savefig('GP Predictor.png', dpi=300)
-
plt.show()
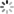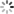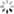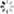
-
# In[11]:随机森林模型
-
rf = RandomForestRegressor(n_estimators=100, oob_score=True, max_features='auto')
-
rf.fit(X_train, y_train)
-
# In[12]:
-
y_pred = rf.predict(X_test)
-
rms = (np.mean((y_test - y_pred)**2))**0.5
-
print ("RF RMS", rms)
-
# out[12]:
-
RF RMS 0.6057144333891424
-
# In[13]:
-
print ("RF r^2 score",r2_score(y_test,y_pred))
-
# out[13]:
-
RF r^2 score 0.9120696293757707
-
# In[14]:结果绘图
-
plt.scatter(y_train,rf.predict(X_train), label = 'Train', c='blue')
-
plt.title('RF Predictor')
-
plt.xlabel('Measured Solubility')
-
plt.ylabel('Predicted Solubility')
-
plt.scatter(y_test,rf.predict(X_test),c='lightgreen', label='Test', alpha = 0.8)
-
plt.legend(loc=4)
-
plt.savefig('RF Predictor.png', dpi=300)
-
plt.show()
文章来源: drugai.blog.csdn.net,作者:DrugAI,版权归原作者所有,如需转载,请联系作者。
原文链接:drugai.blog.csdn.net/article/details/105683682
【版权声明】本文为华为云社区用户转载文章,如果您发现本社区中有涉嫌抄袭的内容,欢迎发送邮件进行举报,并提供相关证据,一经查实,本社区将立刻删除涉嫌侵权内容,举报邮箱:
cloudbbs@huaweicloud.com
- 点赞
- 收藏
- 关注作者

评论(0)