基于机器学习的临床决策支持-ANN
【摘要】
声明:本文示例来自于GitHub用户vkasojhaa的项目,一切权利归其所有,此处仅是自己学习分享。
实现了基于机器学习的乳腺癌的恶性和良性预测,比较了不同机器学习算法之间的性能。主要目的是评估在每种算法的准确性和效率方面对数据进行分类的正确性。
loss
# 损失值:预估值与实际值之间的均方差
optimizer
# 优化器
trainer = ...
声明:本文示例来自于GitHub用户vkasojhaa的项目,一切权利归其所有,此处仅是自己学习分享。
实现了基于机器学习的乳腺癌的恶性和良性预测,比较了不同机器学习算法之间的性能。主要目的是评估在每种算法的准确性和效率方面对数据进行分类的正确性。
loss
# 损失值:预估值与实际值之间的均方差
optimizer
# 优化器
trainer = optimizer.minimize(loss)
# 训练:最小化损失函数
基于机器学习(ANN)的乳腺癌预测
代码示例
-
#导入依赖库
-
#!/usr/bin/python3
-
from __future__ import print_function
-
import keras
-
from keras.datasets import mnist
-
from keras.models import Sequential
-
from keras.layers import Dense, Dropout, Flatten
-
from keras.layers import Conv2D, MaxPooling2D
-
from keras import backend as K
-
import numpy as np
-
import pandas as pd
-
from matplotlib import pyplot as plt
-
from IPython.display import clear_output
-
from keras.utils import plot_model
-
#载入数据并进行数据预处理
-
data = pd.read_csv("data.csv")
-
data.head()
-
data.drop('id',axis=1,inplace=True)
-
data.drop('Unnamed: 32',axis=1,inplace=True)
-
data['diagnosis'] = data['diagnosis'].map({'M':1,'B':0})
-
data.head()
print("Row, Col", data.shape)# (row,col)
(data['diagnosis'][:398]==1).sum(),(data['diagnosis'][:398]==0).sum()
(data['diagnosis'][398:]==1).sum(),(data['diagnosis'][398:]==0).sum()
-
mat=data.as_matrix()
-
mat.shape
-
mat
模型训练:
-
#Using 2 Sigmoid Layers and RMSprop optimizer
-
model2 = Sequential()
-
model2.add(Dense(500, activation='sigmoid', use_bias=True, input_shape=(30,)))
-
model2.add(Dense(1, activation='sigmoid'))
-
keras.optimizers.RMSprop(lr=0.01, rho=0.9, epsilon=None, decay=0.0)
-
model2.compile(optimizer='rmsprop',loss='binary_crossentropy', metrics=['accuracy'])
-
history01 = model2.fit(mat[:,1:],mat[:,0], validation_split=0.3,shuffle=False,epochs=3000, batch_size=128,verbose=0)
-
score = model2.evaluate(mat[:398,1:],mat[:398,0], verbose=0, batch_size=128)
-
print('Train loss:', score[0])
-
print('Train accuracy:', score[1])
-
Train loss: 0.00821199454791287
-
Train accuracy: 1.0
-
score = model2.evaluate(mat[398:,1:],mat[398:,0], verbose=0, batch_size=128)
-
print('Validation loss:', score[0])
-
print('Validation accuracy:', score[1])
-
Validation loss: 0.1262894694568121
-
Validation accuracy: 0.9473684154755888
-
plt.plot(history01.history['acc'], label='acc')
-
plt.plot(history01.history['val_acc'], label='val_acc')
-
plt.legend()
-
plt.show()
-
plt.plot(history01.history['loss'], label='loss')
-
plt.plot(history01.history['val_loss'], label='val_loss')
-
plt.legend()
-
plt.show()
-
count=0
-
for i in history01.history['acc']:
-
if i>0.99:
-
count+=1
-
print(count)
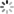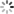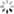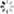
-
#Using 3 Sigmoid Layers and RMSprop optimizer
-
model3 = Sequential()
-
model3.add(Dense(500, activation='sigmoid', use_bias=True, input_shape=(30,)))
-
model3.add(Dense(500, activation='sigmoid', use_bias=True))
-
model3.add(Dense(1, activation='sigmoid'))
-
keras.optimizers.RMSprop(lr=0.01, rho=0.9, epsilon=None, decay=0.0)
-
model3.compile(optimizer='rmsprop',
-
loss='binary_crossentropy',
-
metrics=['accuracy'])
-
history0 = model3.fit(mat[:,1:],mat[:,0], validation_split=0.3,shuffle=False,epochs=3000, batch_size=128, verbose=0)
-
score2 = model3.evaluate(mat[:398,1:],mat[:398,0], verbose=0, batch_size=128)
-
print('Train loss:', score2[0])
-
print('Train accuracy:', score2[1])
-
Train loss: 0.011901359889894986
-
Train accuracy: 0.992462311557789
-
score2 = model3.evaluate(mat[398:,1:],mat[398:,0], verbose=0, batch_size=128)
-
print('Validation loss:', score2[0])
-
print('Validation accuracy:', score2[1])
-
Validation loss: 0.0804629116945448
-
Validation accuracy: 0.9707602311296073
-
plt.plot(history0.history['acc'], label='acc')
-
plt.plot(history0.history['val_acc'], label='val_acc')
-
plt.legend()
-
plt.show()
-
plt.plot(history0.history['loss'], label='loss')
-
plt.plot(history0.history['val_loss'], label='val_loss')
-
plt.legend()
-
plt.show()
-
count=0
-
for i in history0.history['acc']:
-
if i>0.99:
-
count+=1
-
print(count)
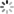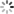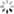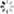
-
#Using 3 Sigmoid Layers and RMSprop optimizer
-
model3 = Sequential()
-
model3.add(Dense(500, activation='sigmoid', use_bias=True, input_shape=(30,)))
-
model3.add(Dense(500, activation='sigmoid', use_bias=True))
-
model3.add(Dense(1, activation='sigmoid'))
-
keras.optimizers.RMSprop(lr=0.01, rho=0.9, epsilon=None, decay=0.0)
-
model3.compile(optimizer='rmsprop',
-
loss='binary_crossentropy',
-
metrics=['accuracy'])
-
history0 = model3.fit(mat[:,1:],mat[:,0], validation_split=0.3,shuffle=False,epochs=3000, batch_size=128, verbose=0)
-
score2 = model3.evaluate(mat[:398,1:],mat[:398,0], verbose=0, batch_size=128)
-
print('Train loss:', score2[0])
-
print('Train accuracy:', score2[1])
-
Validation loss: 0.0804629116945448
-
Validation accuracy: 0.9707602311296073
-
plt.plot(history0.history['acc'], label='acc')
-
plt.plot(history0.history['val_acc'], label='val_acc')
-
plt.legend()
-
plt.show()
-
plt.plot(history0.history['loss'], label='loss')
-
plt.plot(history0.history['val_loss'], label='val_loss')
-
plt.legend()
-
plt.show()
-
count=0
-
for i in history0.history['acc']:
-
if i>0.99:
-
count+=1
-
print(count)
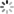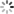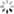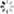
-
#Using 4 Sigmoid Layers and RMSprop optimizer
-
model4 = Sequential()
-
model4.add(Dense(500, activation='sigmoid', use_bias=True, input_shape=(30,)))
-
model4.add(Dense(500, activation='sigmoid', use_bias=True))
-
model4.add(Dense(500, activation='sigmoid', use_bias=True))
-
model4.add(Dense(1, activation='sigmoid'))
-
keras.optimizers.RMSprop(lr=0.01, rho=0.9, epsilon=None, decay=0.0)
-
model4.compile(optimizer='rmsprop', loss='binary_crossentropy', metrics=['accuracy'])
-
history1 = model4.fit(mat[:,1:],mat[:,0], validation_split=0.3,shuffle=False,epochs=3000, batch_size=128, verbose=0)
-
#model.evaluate(mat[:,1:],mat[:,0], batch_size=None, verbose=1, sample_weight=None, steps=None)
-
score = model4.evaluate(mat[:398,1:],mat[:398,0], verbose=0, batch_size=128)
-
print('Train loss:', score[0])
-
print('Train accuracy:', score[1])
-
Train loss: 0.1454299822000403
-
Train accuracy: 0.9346733668341709
-
score = model4.evaluate(mat[398:,1:],mat[398:,0], verbose=0, batch_size=128)
-
print('Validation loss:', score[0])
-
print('Validation accuracy:', score[1])
-
Validation loss: 0.33671002412400053
-
Validation accuracy: 0.8830409339296887
-
plt.plot(history1.history['acc'], label='acc')
-
plt.plot(history1.history['val_acc'], label='val_acc')
-
plt.legend()
-
plt.show()
-
plt.plot(history1.history['loss'], label='loss')
-
plt.plot(history1.history['val_loss'], label='val_loss')
-
plt.legend()
-
plt.show()
-
count=0
-
for i in history1.history['acc']:
-
if i>0.99:
-
count+=1
-
print(count)
Connecting artificial intelligence (AI) with pharmaceutical sciences.
参考资料:
https://github.com/vkasojhaa/Clinical-Decision-Support-using-Machine-Learning
文章来源: drugai.blog.csdn.net,作者:DrugAI,版权归原作者所有,如需转载,请联系作者。
原文链接:drugai.blog.csdn.net/article/details/105683686
【版权声明】本文为华为云社区用户转载文章,如果您发现本社区中有涉嫌抄袭的内容,欢迎发送邮件进行举报,并提供相关证据,一经查实,本社区将立刻删除涉嫌侵权内容,举报邮箱:
cloudbbs@huaweicloud.com
- 点赞
- 收藏
- 关注作者

评论(0)