蛋白序列 | 基于深度学习的蛋白质序列家族分类
背景简介
蛋白质数据集来自于结构生物信息学研究协作组织(RCSB)的蛋白质数据库(PDB)。
RCSB : Research Collaboratory for Structural Bioinformatics
PDB : Protein Data Bank
PDB是原子坐标和描述蛋白质和其他重要生物大分子的信息储存库。结构生物学家使用诸如X射线晶体学、NMR和低温电子显微术的方法来确定分子中每个原子的相对于彼此的位置。
不断发展的PDB反映了世界各地实验室正在进行的研究,使得在研究和教育中使用数据库既令人兴奋又具有挑战性。结构可用于生命过程中涉及的许多蛋白质和核酸,因此可以访问PDB数据库查找核糖体、癌基因、药物靶标甚至整个病毒的结构。但是,找到所需信息可能是一项挑战,因为PDB会存在许多不同的结构,经常发现给定分子或部分结构的多种结构,或已经从其天然形式修饰或失活的结构。
蛋白质序列家族分类
根据氨基酸序列对蛋白质家族进行分类。 工作基于自然语言处理(NLP)中深度学习模型,并假设蛋白质序列可以被视为一种语言。
-
#import library
-
-
import numpy as np
-
import pandas as pd
-
-
from keras.models import Sequential
-
from keras.layers import Dense, Conv1D, MaxPooling1D, Flatten,AtrousConvolution1D,SpatialDropout1D, Dropout
-
from keras.layers.normalization import BatchNormalization
-
-
from keras.layers import LSTM
-
from keras.layers.embeddings import Embedding
-
from keras.callbacks import EarlyStopping
-
from keras.preprocessing import text, sequence
-
from keras.preprocessing.text import Tokenizer
-
from sklearn.model_selection import train_test_split
-
-
%matplotlib inline
-
# Merge the two Data set together
-
# Drop duplicates
-
df = pd.read_csv('pdbdata_no_dups.csv').merge(pd.read_csv('pdbdata_seq.csv'), how='inner', on='structureId').drop_duplicates(["sequence"]) # ,"classification"
-
# Drop rows with missing labels
-
df = df[[type(c) == type('') for c in df.classification.values]]
-
df = df[[type(c) == type('') for c in df.sequence.values]]
-
# select proteins
-
df = df[df.macromoleculeType_x == 'Protein']
-
df.reset_index()
-
print(df.shape)
-
df.head()
(87761, 18)
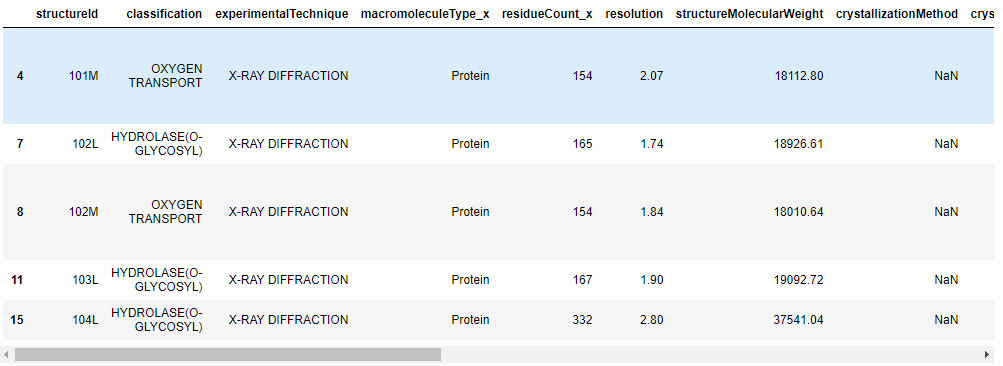
-
print(df.residueCount_x.quantile(0.9))
-
df.residueCount_x.describe()
count 87761.000000 mean 922.913937 std 3173.118920 min 3.000000 25% 234.000000 50% 451.000000 75% 880.000000 max 157478.000000 Name: residueCount_x, dtype: float64
-
df = df.loc[df.residueCount_x<1200]
-
print(df.shape[0])
-
df.residueCount_x.describe()
count 73140.000000 mean 433.599918 std 289.671609 min 3.000000 25% 198.000000 50% 373.000000 75% 618.000000 max 1198.000000 Name: residueCount_x, dtype: float64
EDA
-
import matplotlib.pyplot as plt
-
from collections import Counter
-
-
# count numbers of instances per class
-
cnt = Counter(df.classification)
-
# select only K most common classes! - was 10 by default
-
top_classes = 10
-
# sort classes
-
sorted_classes = cnt.most_common()[:top_classes]
-
classes = [c[0] for c in sorted_classes]
-
counts = [c[1] for c in sorted_classes]
-
print("at least " + str(counts[-1]) + " instances per class")
-
-
# apply to dataframe
-
print(str(df.shape[0]) + " instances before")
-
df = df[[c in classes for c in df.classification]]
-
print(str(df.shape[0]) + " instances after")
-
-
seqs = df.sequence.values
-
lengths = [len(s) for s in seqs]
-
-
# visualize
-
fig, axarr = plt.subplots(1,2, figsize=(20,5))
-
axarr[0].bar(range(len(classes)), counts)
-
plt.sca(axarr[0])
-
plt.xticks(range(len(classes)), classes, rotation='vertical')
-
axarr[0].set_ylabel('frequency')
-
-
axarr[1].hist(lengths, bins=50, normed=False)
-
axarr[1].set_xlabel('sequence length')
-
axarr[1].set_ylabel('# sequences')
-
plt.show()
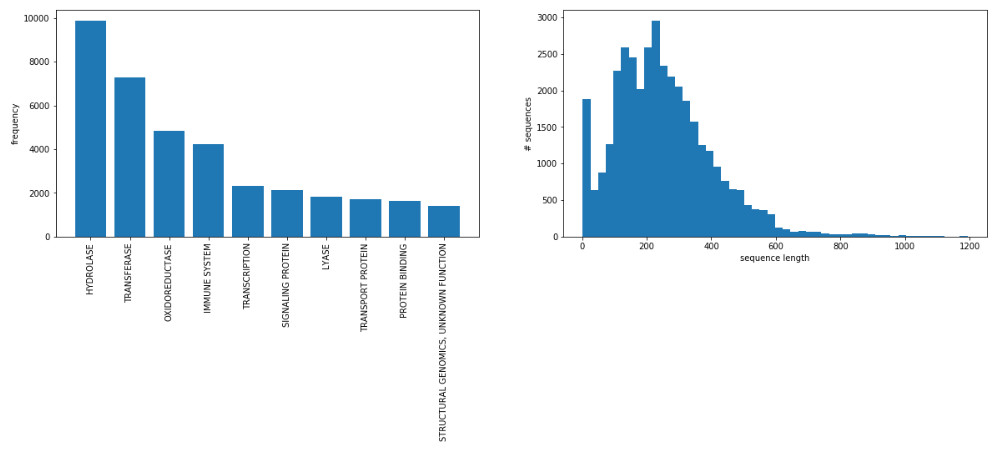
转换标签
首先,数据集被简化为十个最常见类之一的样本。 序列的长度范围从极少数氨基酸到几千个氨基酸。
其次,使用sklearn.preprocessing中的LabelBinarizer将字符串中的标签转换为一个one hot表示。
-
from sklearn.preprocessing import LabelBinarizer
-
-
# Transform labels to one-hot
-
lb = LabelBinarizer()
-
Y = lb.fit_transform(df.classification)
用keras进一步预处理序列
使用keras库进行文本处理:
Tokenizer:将序列的每个字符转换为数字
pad_sequences:确保每个序列具有相同的长度(max_length)。
train_test_split:将数据分成训练和测试样本。
-
# maximum length of sequence, everything afterwards is discarded! Default 256
-
# max_length = 256
-
max_length = 300
-
-
#create and fit tokenizer
-
tokenizer = Tokenizer(char_level=True)
-
tokenizer.fit_on_texts(seqs)
-
#represent input data as word rank number sequences
-
X = tokenizer.texts_to_sequences(seqs)
-
X = sequence.pad_sequences(X, maxlen=max_length)
建立keras模型并适合
NLP中取得的成功建议使用已经实现为keras层(嵌入)的字嵌入。本例中,只有20个不同的单词(每个氨基酸)。可以尝试对序列的one hot表示进行2D卷积,而不是单词嵌入,但是这种方法侧重于将NLP理论应用于蛋白质序列。
为了提高性能,还可以使用深层体系结构(后续的卷积和池化层),这里有两层,其中第一层有64个滤波器,卷积大小为6,第二层有32个滤波器,大小为3。
将激活平展并传递到完全相同的层,其中最后一层是softmax激活,并且大小对应于类的数量。
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=.2)
-
embedding_dim = 11 # orig 8
-
-
# create the model
-
model = Sequential()
-
model.add(Embedding(len(tokenizer.word_index)+1, embedding_dim, input_length=max_length))
-
# model.add(Conv1D(filters=64, kernel_size=6, padding='same', activation='relu')) #orig
-
model.add(Conv1D(filters=128, kernel_size=9, padding='valid', activation='relu',dilation_rate=1))
-
# model.add(BatchNormalization())
-
model.add(MaxPooling1D(pool_size=2))
-
# model.add(Conv1D(filters=32, kernel_size=3, padding='same', activation='relu')) # orig
-
model.add(Conv1D(filters=64, kernel_size=4, padding='valid', activation='relu'))
-
model.add(BatchNormalization())
-
model.add(MaxPooling1D(pool_size=2))
-
model.add(Flatten())
-
model.add(Dense(200, activation='elu')) # 128
-
# model.add(BatchNormalization())
-
# model.add(Dense(64, activation='elu')) # 128
-
model.add(BatchNormalization())
-
model.add(Dense(top_classes, activation='softmax'))
-
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
-
print(model.summary())
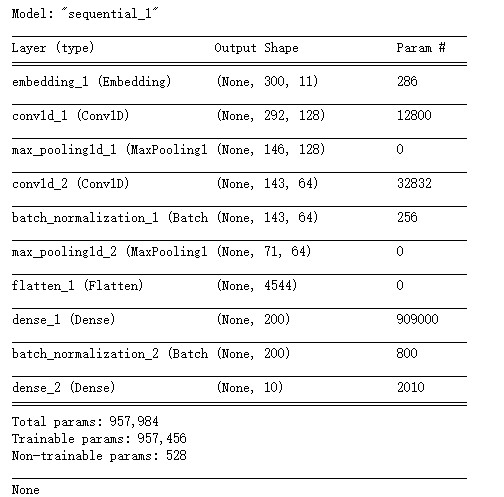
-
es = EarlyStopping(monitor='val_acc', verbose=1, patience=4)
-
-
model.fit(X_train, y_train, batch_size=128, verbose=1, validation_split=0.15,callbacks=[es],epochs=25) # epochs=15, # batch_size=128

-
from keras.layers import Conv1D, MaxPooling1D, Concatenate, Input
-
from keras.models import Sequential,Model
-
-
units = 256
-
num_filters = 32
-
filter_sizes=(3,5, 9,15,21)
-
conv_blocks = []
-
-
embedding_layer = Embedding(len(tokenizer.word_index)+1, embedding_dim, input_length=max_length)
-
es2 = EarlyStopping(monitor='val_acc', verbose=1, patience=4)
-
-
sequence_input = Input(shape=(max_length,), dtype='int32')
-
embedded_sequences = embedding_layer(sequence_input)
-
-
z = Dropout(0.1)(embedded_sequences)
-
-
for sz in filter_sizes:
-
conv = Conv1D(
-
filters=num_filters,
-
kernel_size=sz,
-
padding="valid",
-
activation="relu",
-
strides=1)(z)
-
conv = MaxPooling1D(pool_size=2)(conv)
-
conv = Flatten()(conv)
-
conv_blocks.append(conv)
-
z = Concatenate()(conv_blocks) if len(conv_blocks) > 1 else conv_blocks[0]
-
z = Dropout(0.25)(z)
-
z = BatchNormalization()(z)
-
z = Dense(units, activation="relu")(z)
-
z = BatchNormalization()(z)
-
predictions = Dense(top_classes, activation="softmax")(z)
-
model2 = Model(sequence_input, predictions)
-
model2.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
-
print(model2.summary())
-
-
model2.fit(X_train, y_train, batch_size=64, verbose=1, validation_split=0.15,callbacks=[es],epochs=30)
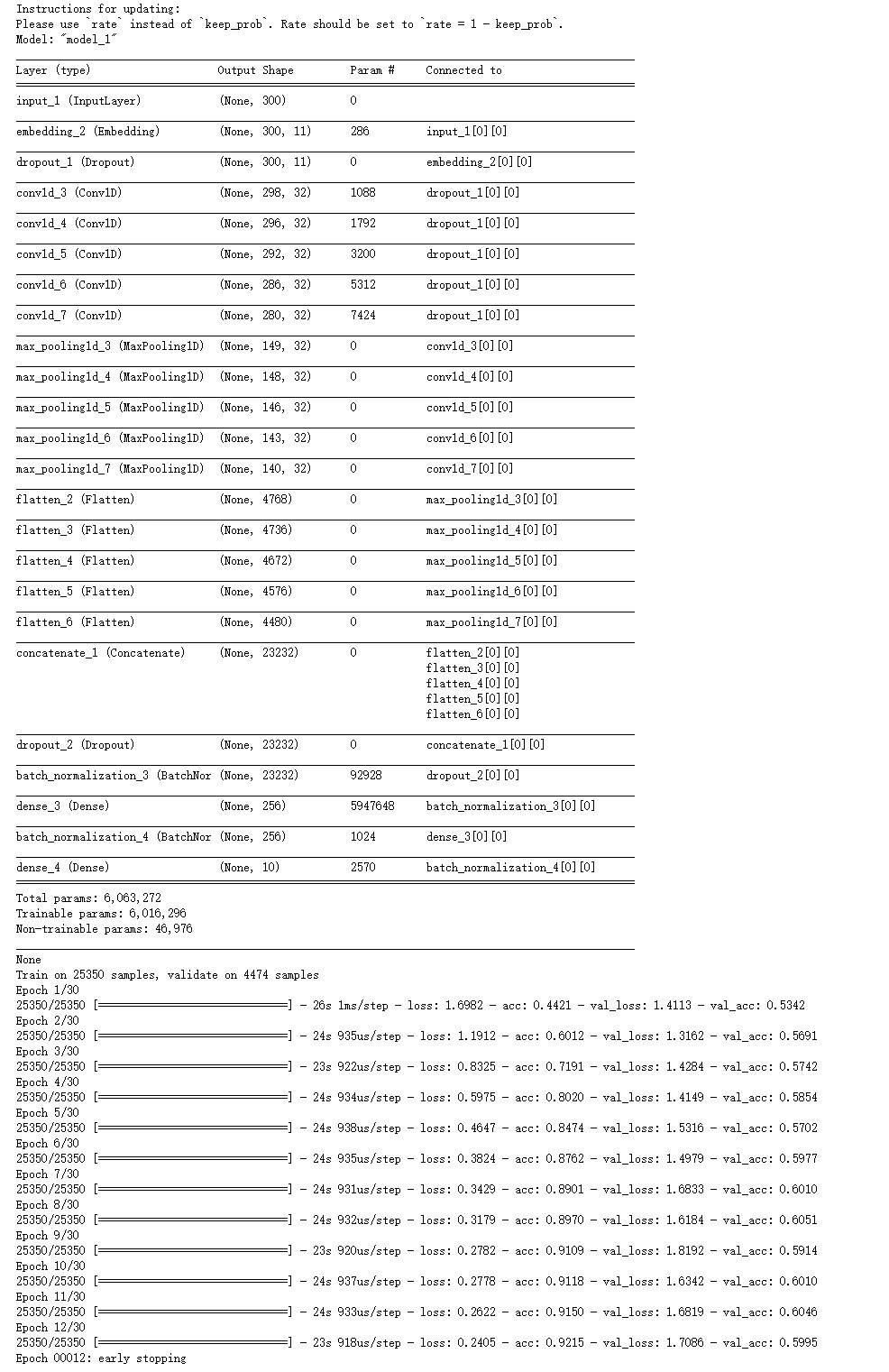
-
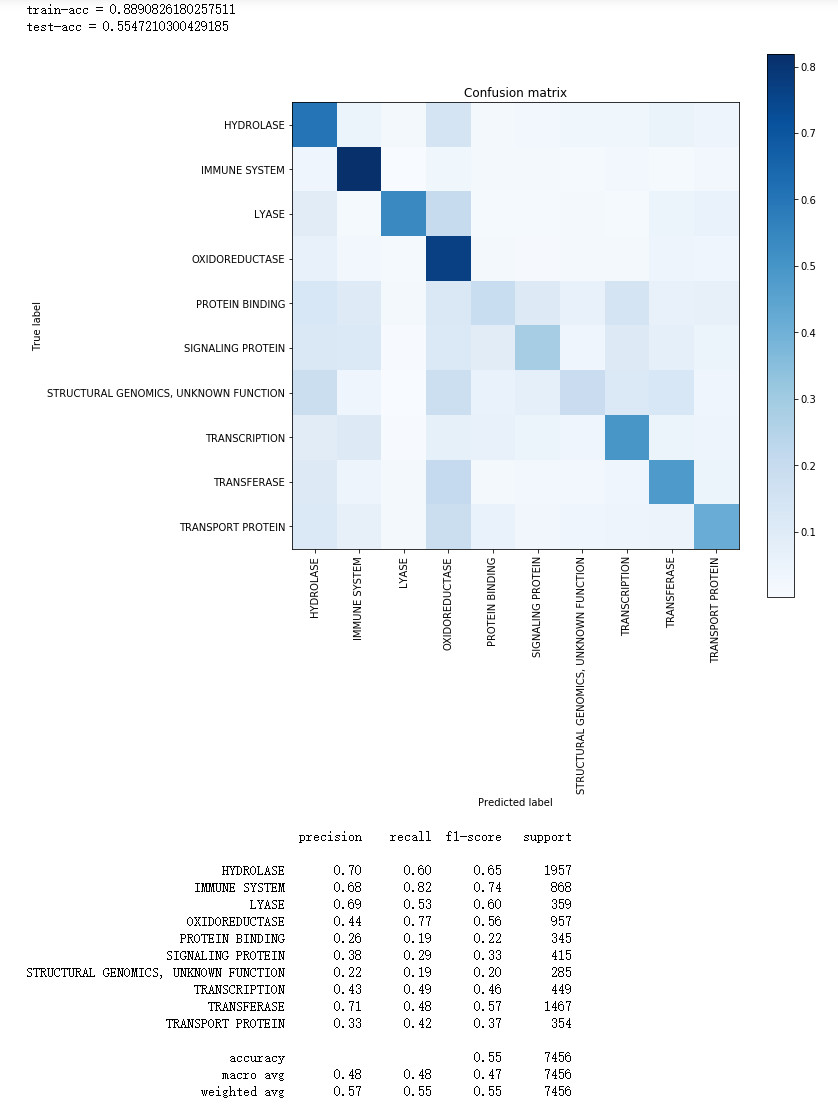
from sklearn.metrics import confusion_matrix
-
from sklearn.metrics import accuracy_score
-
from sklearn.metrics import classification_report
-
import itertools
-
-
train_pred = model.predict(X_train)
-
test_pred = model.predict(X_test)
-
print("train-acc = " + str(accuracy_score(np.argmax(y_train, axis=1), np.argmax(train_pred, axis=1))))
-
print("test-acc = " + str(accuracy_score(np.argmax(y_test, axis=1), np.argmax(test_pred, axis=1))))
-
-
# Compute confusion matrix
-
cm = confusion_matrix(np.argmax(y_test, axis=1), np.argmax(test_pred, axis=1))
-
-
# Plot normalized confusion matrix
-
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
-
np.set_printoptions(precision=2)
-
plt.figure(figsize=(10,10))
-
plt.imshow(cm, interpolation='nearest', cmap=plt.cm.Blues)
-
plt.title('Confusion matrix')
-
plt.colorbar()
-
tick_marks = np.arange(len(lb.classes_))
-
plt.xticks(tick_marks, lb.classes_, rotation=90)
-
plt.yticks(tick_marks, lb.classes_)
-
#for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
-
# plt.text(j, i, format(cm[i, j], '.2f'), horizontalalignment="center", color="white" if cm[i, j] > cm.max() / 2. else "black")
-
plt.ylabel('True label')
-
plt.xlabel('Predicted label')
-
plt.show()
-
-
print(classification_report(np.argmax(y_test, axis=1), np.argmax(test_pred, axis=1), target_names=lb.classes_))
-
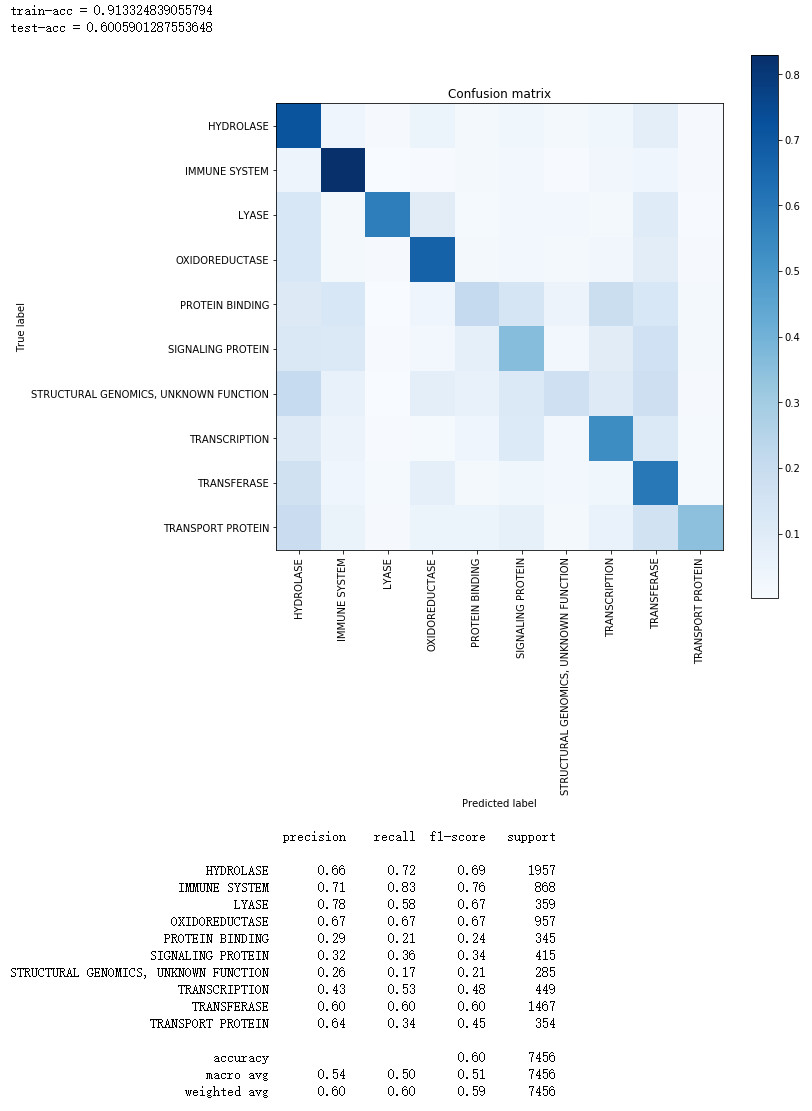
train_pred = model2.predict(X_train)
-
test_pred = model2.predict(X_test)
-
print("train-acc = " + str(accuracy_score(np.argmax(y_train, axis=1), np.argmax(train_pred, axis=1))))
-
print("test-acc = " + str(accuracy_score(np.argmax(y_test, axis=1), np.argmax(test_pred, axis=1))))
-
-
# Compute confusion matrix
-
cm = confusion_matrix(np.argmax(y_test, axis=1), np.argmax(test_pred, axis=1))
-
-
# Plot normalized confusion matrix
-
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
-
np.set_printoptions(precision=2)
-
plt.figure(figsize=(10,10))
-
plt.imshow(cm, interpolation='nearest', cmap=plt.cm.Blues)
-
plt.title('Confusion matrix')
-
plt.colorbar()
-
tick_marks = np.arange(len(lb.classes_))
-
plt.xticks(tick_marks, lb.classes_, rotation=90)
-
plt.yticks(tick_marks, lb.classes_)
-
#for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
-
# plt.text(j, i, format(cm[i, j], '.2f'), horizontalalignment="center", color="white" if cm[i, j] > cm.max() / 2. else "black")
-
plt.ylabel('True label')
-
plt.xlabel('Predicted label')
-
plt.show()
-
-
print(classification_report(np.argmax(y_test, axis=1), np.argmax(test_pred, axis=1), target_names=lb.classes_))
参考资料:
文章来源: drugai.blog.csdn.net,作者:DrugAI,版权归原作者所有,如需转载,请联系作者。
原文链接:drugai.blog.csdn.net/article/details/100690926
- 点赞
- 收藏
- 关注作者

评论(0)